Articles
- Page Path
- HOME > J Mov Disord > Volume 13(1); 2020 > Article
-
Original Article
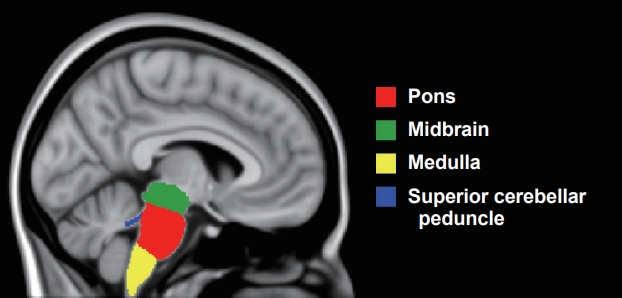
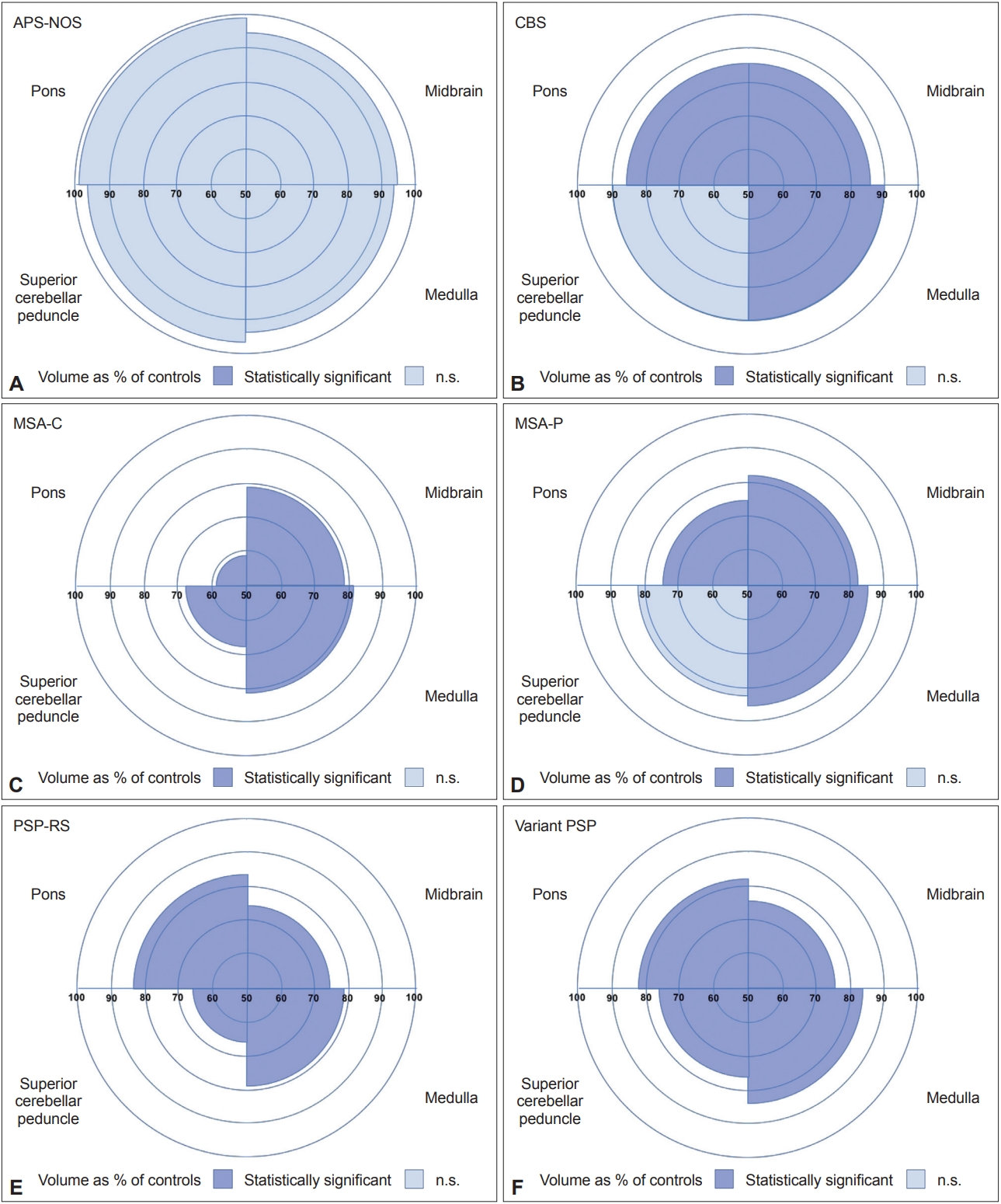
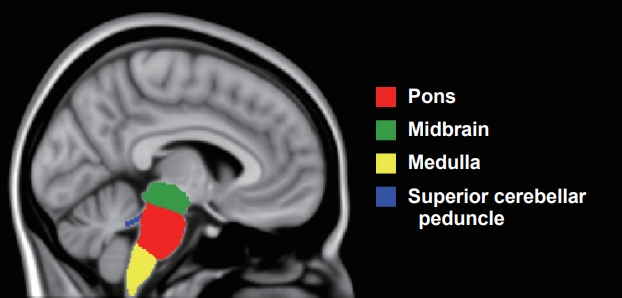
Automated Brainstem Segmentation Detects Differential Involvement in Atypical Parkinsonian Syndromes -
Martina Bocchetta1
 , Juan Eugenio Iglesias2, Viorica Chelban3,4, Edwin Jabbari3, Ruth Lamb3, Lucy L. Russell1, Caroline V. Greaves1, Mollie Neason1, David M. Cash1,2, David L. Thomas5,6, Jason D. Warren1, John Woodside3, Henry Houlden3, Huw R. Morris3, Jonathan D. Rohrer1
, Juan Eugenio Iglesias2, Viorica Chelban3,4, Edwin Jabbari3, Ruth Lamb3, Lucy L. Russell1, Caroline V. Greaves1, Mollie Neason1, David M. Cash1,2, David L. Thomas5,6, Jason D. Warren1, John Woodside3, Henry Houlden3, Huw R. Morris3, Jonathan D. Rohrer1
-
Journal of Movement Disorders 2020;13(1):39-46.
DOI: https://doi.org/10.14802/jmd.19030
Published online: September 26, 2019
1Dementia Research Centre, Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, University College London, London, UK
2Centre for Medical Image Computing, Department of Medical Physics and Biomedical Engineering, University College London, London, UK
3Department of Movement Disorders, UCL Queen Square Institute of Neurology, University College London, London, UK
4Department of Neurology and Neurosurgery, Institute of Emergency Medicine, Chisinau, Republic of Moldova
5Neuroradiological Academic Unit, UCL Queen Square Institute of Neurology, University College London, London, UK
6Leonard Wolfson Experimental Neurology Centre, UCL Queen Square Institute of Neurology, University College London, London, UK
- Corresponding author: Jonathan D. Rohrer, FRCP, PhD Dementia Research Centre, Department of Neurodegenerative Disease, UCL Queen Square Institute of Neurology, University College London, 8-11 Queen Square, London WC1N 3AR, UK / Tel: +44-0-7738271475 / Fax: +44-0-20-34483104 / E-mail: j.rohrer@ucl.ac.uk
Copyright © 2020 The Korean Movement Disorder Society
This is an Open Access article distributed under the terms of the Creative Commons Attribution Non-Commercial License (http://creativecommons.org/licenses/by-nc/4.0/) which permits unrestricted non-commercial use, distribution, and reproduction in any medium, provided the original work is properly cited.
Figure & Data
References
Citations

- Loss of brainstem white matter predicts onset and motor neuron symptoms in C9orf72 expansion carriers: a GENFI study
Agnès Pérez-Millan, Sergi Borrego-Écija, John C. van Swieten, Lize Jiskoot, Fermin Moreno, Robert Laforce, Caroline Graff, Mario Masellis, Maria Carmela Tartaglia, James B. Rowe, Barbara Borroni, Elizabeth Finger, Matthis Synofzik, Daniela Galimberti, Rik
Journal of Neurology.2023; 270(3): 1573. CrossRef - Comparative validation of AI and non-AI methods in MRI volumetry to diagnose Parkinsonian syndromes
Joomee Song, Juyoung Hahm, Jisoo Lee, Chae Yeon Lim, Myung Jin Chung, Jinyoung Youn, Jin Whan Cho, Jong Hyeon Ahn, Kyungsu Kim
Scientific Reports.2023;[Epub] CrossRef - Identification of the pathogenesis features of various phenotypes of multiple sclerosis based on the study of the morphological functional connectivity of subcortical gray matter structures
A. G. Trufanov, A. Y. Polushin, E. A. Gorbunova, M. V. Lukin
Russian Journal for Personalized Medicine.2023; 3(1): 27. CrossRef - Structural MRI predicts clinical progression in presymptomatic genetic frontotemporal dementia: findings from the GENetic Frontotemporal dementia Initiative cohort
Martina Bocchetta, Emily G Todd, Arabella Bouzigues, David M Cash, Jennifer M Nicholas, Rhian S Convery, Lucy L Russell, David L Thomas, Ian B Malone, Juan Eugenio Iglesias, John C van Swieten, Lize C Jiskoot, Harro Seelaar, Barbara Borroni, Daniela Galim
Brain Communications.2023;[Epub] CrossRef - Quantitative MRI protocol and decision model for a ‘one stop shop’ early-stage Parkinsonism diagnosis: Study design
Samy Abo Seada, Anke W. van der Eerden, Agnita J.W. Boon, Juan A. Hernandez-Tamames
NeuroImage: Clinical.2023; 39: 103506. CrossRef - Neuroimaging correlates of brain injury in Wilson’s disease: a multimodal, whole-brain MRI study
Samuel Shribman, Martina Bocchetta, Carole H Sudre, Julio Acosta-Cabronero, Maggie Burrows, Paul Cook, David L Thomas, Godfrey T Gillett, Emmanuel A Tsochatzis, Oliver Bandmann, Jonathan D Rohrer, Thomas T Warner
Brain.2022; 145(1): 263. CrossRef - Nuclear imaging in Parkinson's disease: The past, the present, and the future
Haim Golan, Olga Volkov, Eli Shalom
Journal of the Neurological Sciences.2022; 436: 120220. CrossRef - Criteria-unfulfilled multiple system atrophy at an initial stage exhibits laterality of middle cerebellar peduncles
Minori Furuta, Masayuki Sato, Setsuki Tsukagoshi, Yoshito Tsushima, Yoshio Ikeda
Journal of the Neurological Sciences.2022; 438: 120281. CrossRef - A data-driven model of brain volume changes in progressive supranuclear palsy
W. J. Scotton, M. Bocchetta, E. Todd, D. M. Cash, N. Oxtoby, L. VandeVrede, H. Heuer, D. C. Alexander, J. B. Rowe, H. R. Morris, A. Boxer, J. D. Rohrer, P. A. Wijeratne
Brain Communications.2022;[Epub] CrossRef - Neuroimaging Correlates of Cognitive Deficits in Wilson's Disease
Samuel Shribman, Maggie Burrows, Rhian Convery, Martina Bocchetta, Carole H. Sudre, Julio Acosta‐Cabronero, David L. Thomas, Godfrey T. Gillett, Emmanuel A. Tsochatzis, Oliver Bandmann, Jonathan D. Rohrer, Thomas T. Warner
Movement Disorders.2022; 37(8): 1728. CrossRef - Corticobasal syndrome and Parkinson’s disease at the beginning: asymmetrical patterns of MRI and Blink Reflex for early diagnosis
Giulia Donzuso, Giorgia Sciacca, Antonina Luca, Calogero E. Cicero, Giovanni Mostile, Alessandra Nicoletti, Mario Zappia
Journal of Neural Transmission.2022; 129(12): 1427. CrossRef - Eye movements and association with regional brain atrophy in clinical subtypes of progressive supranuclear palsy
Ji-Hyun Choi, Heejung Kim, Jung Hwan Shin, Jee-Young Lee, Han-Joon Kim, Jong-Min Kim, Beomseok Jeon
Journal of Neurology.2021; 268(3): 967. CrossRef - Artificial intelligence applied to neuroimaging data in Parkinsonian syndromes: Actuality and expectations
Annalisa Vitale, Rossella Villa, Lorenzo Ugga, Valeria Romeo, Arnaldo Stanzione, Renato Cuocolo
Mathematical Biosciences and Engineering.2021; 18(2): 1753. CrossRef - Differential early subcortical involvement in genetic FTD within the GENFI cohort
Martina Bocchetta, Emily G. Todd, Georgia Peakman, David M. Cash, Rhian S. Convery, Lucy L. Russell, David L. Thomas, Juan Eugenio Iglesias, John C. van Swieten, Lize C. Jiskoot, Harro Seelaar, Barbara Borroni, Daniela Galimberti, Raquel Sanchez-Valle, Ro
NeuroImage: Clinical.2021; 30: 102646. CrossRef - Quantitative MRI markers in Parkinson's disease and parkinsonian syndromes
Germain Arribarat, Patrice Péran
Current Opinion in Neurology.2020; 33(2): 222. CrossRef
Comments on this article
 KMDS
KMDS
 E-submission
E-submission
 PubReader
PubReader ePub Link
ePub Link Cite
Cite